GRÜNWALD LAB
Tumour Heterogeneity & Translational Systems Biology
We apply a systems biology approach to understand the molecular mechanisms that govern tumor spatial diversity and to provide actionable tools to clinically navigate this phenomenon. To this end, we often combine traditional methods such as histopathology with modern multimodal profiling setups, applied to human tissues and patient-derived disease models.
Below is a brief description of the current research priorities in the lab.

Univ.-Prof. Dr. rer. nat.
Barbara Grünwald
Head of Experimental Urology. Research Group Leader: Translational Systems Biology
Tissue self-organization in solid tumors
A major focus in the lab lies on pancreatic ductal adenocarcinoma (PDAC), which remains one of the most challenging cancers to treat. PDAC is a notoriously heterogeneous cancer type, characterized by a complex tumor microenvironment (TME) which has complicated mechanistic and preclinical studies for decades. This heterogeneity was long thought to be the result of random cell changes representative of global tumor disorganization. Based on our own findings and a growing body of evidence in the literature, we are convinced that heterogeneity in solid tumors is not at all random but a critical foundation of malignant tissue function and adaptive behavior. In fact, solid tumors exhibit a considerable level of functional self-organization that we have barely appreciated. For instance, in PDAC, we find distinct spatial programs that are vastly distinct from each other, yet, highly conserved between different patients and disease sites. Their intratumoral co-existence instigates massive spatial heterogeneity yet, this heterogeneity is a well-definable function of few, fundamental tissue organizational mechanisms. In other words, despite their famous heterogeneity, PDAC tissues facto self-organize into relatively few states of functional relevance. Ongoing efforts in the lab are to understand whether other solid tumor types (lung, kidney, bladder) also utilize such spatial self-organizational programs, what their organ-specific tumor relevant functions are, and how we can diagnostically access this information to aid clinical decision making.
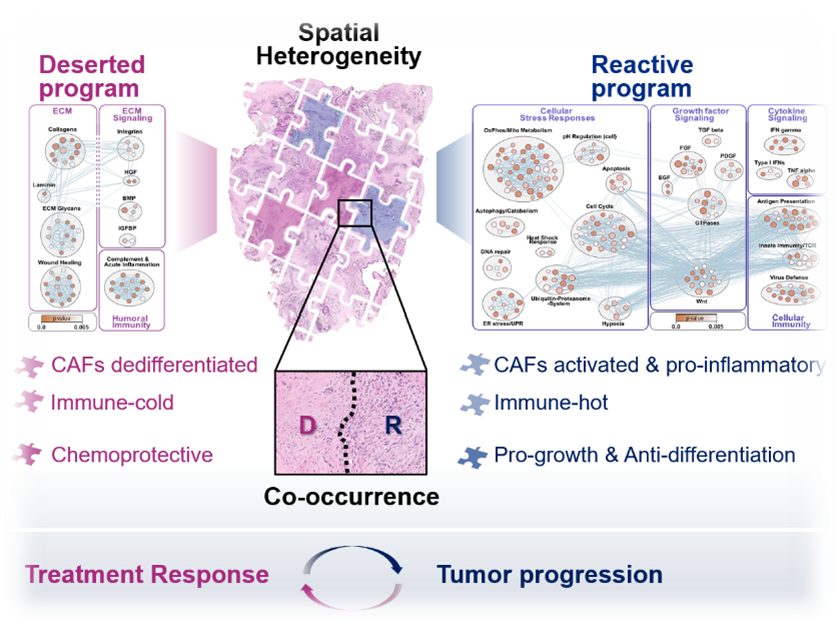
Drivers of tumoral heterogeneity: Tumor-TME-Interplay
The heterogeneous cellular landscape of tumors provides a battery of unique habitats that influence cancer cell phenotypes and behavior, treatment response, as well as immune functions. Understanding the mechanisms that drive this fundamental property of tumor tissues is essential for developing more effective cancer treatments. We are pairing deep tissue profiling with self-organizing human model systems to identify and target the molecular pathways that shape major tissue states within the heterogenous tumor landscape. In addition to macroscopic organizational programs, we are exploring how the intimate interplay between specific malignant and non-malignant cell populations in the tumor shapes the emergence of clinically relevant epithelial tumor subtypes.
Ecosystem co-evolution in cancer initiation
At the earliest stages of tumor initiation, neoplastic cells not only have to acquire the relevant driver mutations but also must successfully interact with an ever-evolving microenvironment. In this interaction, loss of tissue organization is a key permissive event in malignant transformation. Thorough understanding of the tumor ecosystem during transformation, beyond targeted analysis of the malignant epithelium alone, can thus provide major mechanistic insight and open much-needed diagnostic opportunities for accurate and longitudinal risk assessment.
In pursuit of earlier diagnosis and effective secondary prevention strategies as key unmet needs to improve patient outcomes, we are addressing the mechanisms of ecosystem co-evolution in pancreatic cancer initiation.
Clinical heterogeneity to mechanisms
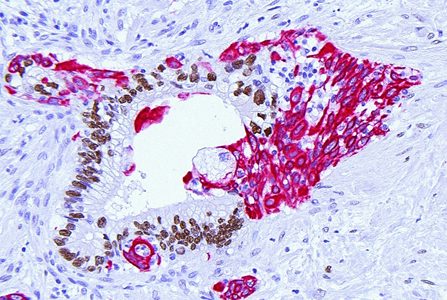
Tumors grow in people and every person is different. Why do some patients respond to a treatment while others don’t? The study of ultra-large cohorts (> 500.000 individuals) has outlined a considerable impact of confounding factors such as age, ethnic background, lifestyle, environmental exposures, and sex on the outcomes of genetic analyses and therapeutic targeting. Such ‘clinical heterogeneity’ has major implications for the design of mechanistic, experimental studies that are aimed at actual clinical progress. For instance, investigating effects of IL-6 in PDAC, we queried any potential confounders and found that the observed IL-6 driven immune phenotypes were consistent across transcriptional subtypes but sex-specific (https://pubmed.ncbi.nlm.nih.gov/37768107/). Beyond systematically testing such confounders in all our research questions, we are now specifically pursuing sexual dimorphisms in major pathways driving intratumoral immune heterogeneity. Furthermore, we are leveraging detailed epidemiological data to fully understand the impact of risk factors on the tumor biology of the resultant tumors.
Heterogeneity-informed biomarkers
Understanding tissue heterogeneity and the underlying biology can help improve clinical practice. While clinical translation of basic research efforts is a major challenge, we are convinced it must be the goal. By design, our deep tissue profiling approaches are firmly anchored in human histopathology, which facilitates a comprehensive portrayal of the functional tumor landscape through a clinically accessible lens. Likewise, transcriptional subtypes hold potential to guide personalized treatment decisions and clinical predictions, yet transcriptomic profiling is not clinically accessible as a routine diagnostic test. Plus, most transcriptional subtypes are not discrete global states but co-occur intratumorally, demanding tissue based diagnostic tests. Through close collaborations with our partners from oncology and pathology, we are condensing insights from global profiling (transcriptomics, integrated multiOMICs) into heterogeneity-informed biomarker assays suitable for prospective validation of the observed associations with treatment response and prognosis.

Multimodal Tissue Phenotyping
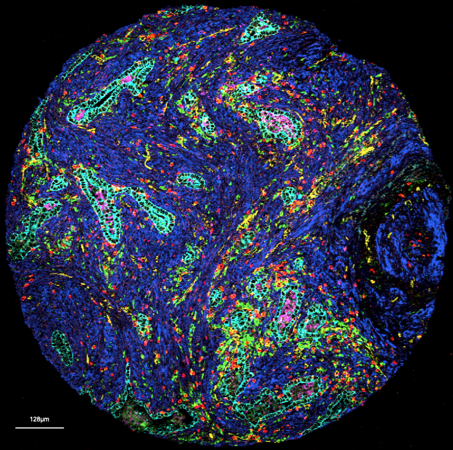
Histopathology remains a bedrock of our approaches and subsequent analyses. Histology reflects tissue organization and we have capitalized on this conserved form-and-function relationship in many of our analytical approaches. Beyond traditional histopathology we are now increasingly using deep learning models to extract relevant histotypes and resultant larger scale topologies from H&Es. For subsequent profiling of defined histological phenotypes, we employ various histology-guided spatial profiling techniques such as Laser Capture Microdissection coupled to multiOMICs, CODEX and Imaging Mass Cytometry. Our wide analytical portfolio is enabled through phenomenal, long-standing collaborators active in technology development who continue to make their newest techniques available to our research questions.
Human 3D model systems
We have assembled a solid collection of patient-derived tumor models as well as stromal cell cultures in our lab and are continuously growing this living biobank together with our collaborators. These three-dimensional organoid models allow us to mimic some of the complexities of tumors as well as different tumor subtypes in vitro and offer a unique platform for studying disease progression and therapeutic responses. We have previously established various Cancer Associated Fibroblast (CAFs) lines from primary patient tumors that recapitulate histomorphological phenotypes of their different originating tissue regions. In heterotypic cultures of tumor organoids, fibroblasts and immune cells, we model the complex, spatial interplay between tumor and TME cells in a controlled manner. Our new Cytation5 Live Cell Imager allows us to record longitudinal parallel morphometric analyses, drug/interference assays and cell type-specific imaging in high-throughput formats. We are developing these heterogeneity-informed platforms to learn about mechanisms of how the tumor-TME interaction shapes malignant behavior but also to understand differences in drug response in a patient specific manner.